The Unmatched Sub-family includes Unmatched surface proteins.
To have direct access to each entry in Unmatched main family, click on its uniprot id on the left bar.
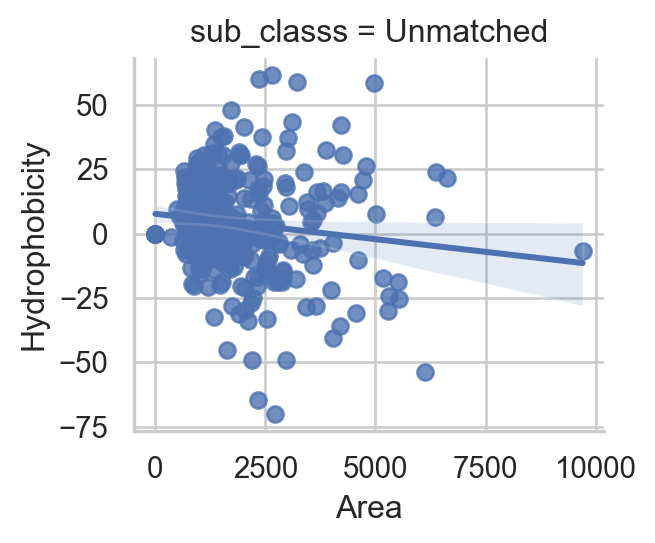
Binding site feature correlation
Code
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set_theme(style="whitegrid")
df_total_flat = pd.read_csv('../database/df_flattened.csv')
unmatched_df1 = df_total_flat.loc[df_total_flat['main_classs'] == "Unmatched"]
ax = sns.lmplot(
data=unmatched_df1, x="areass", y="hpss",
hue="sub_classs", col="sub_classs", height=3, col_wrap=3,
)
ax.set(xlabel ="Area", ylabel = "Hydrophobicity")
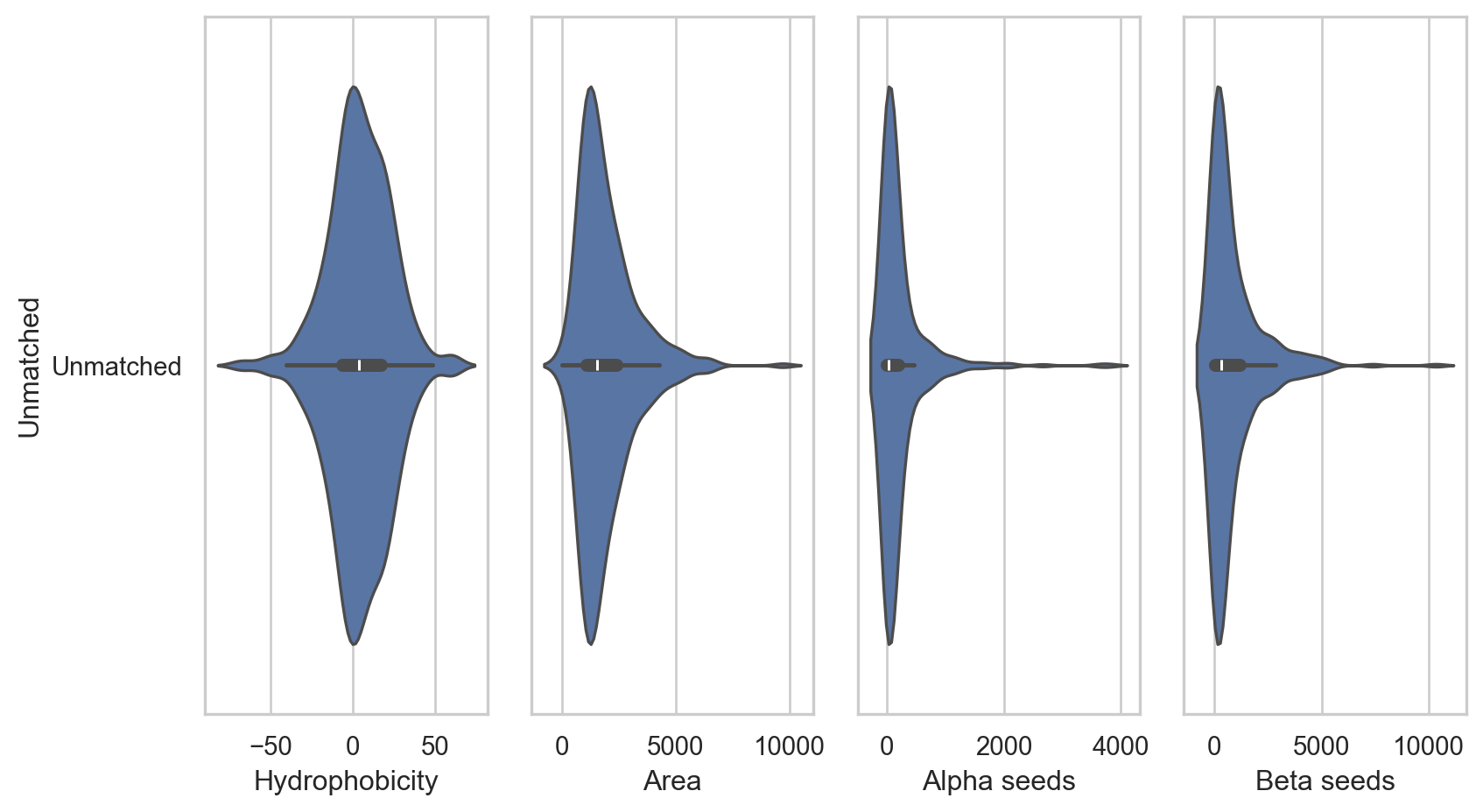
Binding site feature distribution
Code
fig, axes = plt.subplots(1, 4, figsize=(9, 5))
sns.violinplot(
y=unmatched_df1['sub_classs'],
x=unmatched_df1['hpss'],
hue=unmatched_df1['sub_classs'],
ax=axes[0]
)
axes[0].set_xlabel('Hydrophobicity')
axes[0].set_ylabel('Unmatched')
sns.violinplot(
y=unmatched_df1['sub_classs'],
x=unmatched_df1['areass'],
hue=unmatched_df1['sub_classs'],
ax=axes[1]
)
axes[1].set(yticklabels=[])
axes[1].set_ylabel('')
axes[1].set_xlabel('Area')
sns.violinplot(
y=unmatched_df1['sub_classs'],
x=unmatched_df1['seedss_a'],
hue=unmatched_df1['sub_classs'],
ax=axes[2]
)
axes[2].set(yticklabels=[])
axes[2].set_ylabel('')
axes[2].set_xlabel('Alpha seeds')
sns.violinplot(
y=unmatched_df1['sub_classs'],
x=unmatched_df1['seedss_b'],
hue=unmatched_df1['sub_classs'],
ax=axes[3]
)
axes[3].set(yticklabels=[])
axes[3].set_ylabel('')
axes[3].set_xlabel('Beta seeds')
plt.tight_layout()
plt.show()